This article describes the architecture of the Scala collections framework in detail. Compared to the the Scala 2.8 Collections API article, you will find out more about the internal workings of the framework. You will also learn how this architecture helps you define your own collections in a few lines of code, while reusing the overwhelming part of collection functionality from the framework.
The Scala 2.8 Collections API contains a large number of collection operations, which exist uniformly on many different collection implementations. Implementing every collection operation anew for every collection type would lead to an enormous amount of code, most of which would be copied from somewhere else. Such code duplication could lead to inconsistencies over time, when an operation is added or modified in one part of the collection library but not in others. The principal design objective of the new collections framework was to avoid any duplication, defining every operation in as few places as possible. (Ideally, everything should be defined in one place only, but there are a few exceptions where things needed to be redefined.) The design approach was to implement most operations in collection "templates" that can be flexibly inherited from individual base classes and implementations. This article explains these templates and other classes and traits that constitute the "building blocks" of the framework, as well as the construction principles they support.
Builders
| package scala.collection.generic | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| class Builder[-Elem, +To] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def +=(elem: Elem): this.type | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def result(): To | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def clear() | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def mapResult(f: To => NewTo): Builder[Elem, NewTo] = ... | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Almost all collection operations are implemented in terms of traversals and builders. Traversals are handled by Traversable's foreach method, and building new collections is handled by instances of class Builder. Listing 1 presents a slightly abbreviated outline of this class.
You can add an element x to a builder b with b += x. There's also syntax to add more than one element at once, for instance b += (x, y), and b ++= xs work as for buffers (in fact, buffers are an enriched version of builders). The result() method returns a collection from a builder. The state of the builder is undefined after taking its result, but it can be reset into a new empty state using clear(). Builders are generic in both the element type, Elem, and in the type, To, of collections they return.
Often, a builder can refer to some other builder for assembling the elements of a collection, but then would like to transform the result of the other builder, for example to give it a different type. This task is simplified by method mapResult in class Builder. Suppose for instance you have an array buffer buf. Array buffers are builders for themselves, so taking the result() of an array buffer will return the same buffer. If you want to use this buffer to produce a builder that builds arrays, you can use mapResult like this:
| scala> val buf = new ArrayBuffer[Int] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| buf: scala.collection.mutable.ArrayBuffer[Int] = ArrayBuffer() | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> val bldr = buf mapResult (_.toArray) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| bldr: scala.collection.mutable.Builder[Int,Array[Int]] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| = ArrayBuffer() |
Factoring out common operations
| package scala.collection | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| class TraversableLike[+Elem, +Repr] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def newBuilder: Builder[Elem, Repr] // deferred | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def foreach[U](f: Elem => U) // deferred | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ... | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def filter(p: Elem => Boolean): Repr = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val b = newBuilder | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| foreach { elem => if (p(elem)) b += elem } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| b.result | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
The main design objectives of the collection library redesign were to have, at the same time, natural types and maximal sharing of implementation code. In particular, Scala's collections follow the "same-result-type" principle: wherever possible, a transformation method on a collection will yield a collection of the same type. For instance, the filter operation should yield, on every collection type, an instance of the same collection type. Applying filter on a List should give a List; applying it on a Map should give a Map, and so on. In the rest of this section, you will find out how this is achieved.
The Scala collection library avoids code duplication and achieves the "same-result-type" principle by using generic builders and traversals over collections in so-called implementation traits. These traits are named with a Like suffix; for instance, IndexedSeqLike is the implementation trait for IndexedSeq, and similarly, TraversableLike is the implementation trait for Traversable. Collection classes such as Traversable or IndexedSeq inherit all their concrete method implementations from these traits. Implementation traits have two type parameters instead of one for normal collections. They parameterize not only over the collection's element type, but also over the collection's representation type, i.e., the type of the underlying collection, such as Seq[I] or List[T]. For instance, here is the header of trait TraversableLike:
| trait TraversableLike[+Elem, +Repr] { ... } |
Taking filter as an example, this operation is defined once for all collection classes in the trait TraversableLike. An outline of the relevant code is shown in Listing 2. The trait declares two abstract methods, newBuilder and foreach, which are implemented in concrete collection classes. The filter operation is implemented in the same way for all collections using these methods. It first constructs a new builder for the representation type Repr, using newBuilder. It then traverses all elements of the current collection, using foreach. If an element x satisfies the given predicate p (i.e., p(x) is true), it is added with the builder. Finally, the elements collected in the builder are returned as an instance of the Repr collection type by calling the builder's result method.
A bit more complicated is the map operation on collections. For instance, if f is a function from String to Int, and xs is a List[String], then xs map f should give a List[Int]. Likewise, if ys is an Array[String], then ys map f should give a Array[Int]. The problem is how to achieve that without duplicating the definition of the map method in lists and arrays. The newBuilder/foreach framework shown in class TraversableLike, in Listing 2, is not sufficient for this because it only allows creation of new instances of the same collection type whereas map needs an instance of the same collection type constructor, but possibly with a different element type.
What's more, even the result type constructor of a function like map might depend in non-trivial ways on the other argument types. Here is an example:
| scala> import collection.immutable.BitSet | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import collection.immutable.BitSet | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> val bits = BitSet(1, 2, 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| bits: scala.collection.immutable.BitSet = BitSet(1, 2, 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> bits map (_ * 2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res13: scala.collection.immutable.BitSet = BitSet(2, 4, 6) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> bits map (_.toFloat) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res14: scala.collection.immutable.Set[Float] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| = Set(1.0, 2.0, 3.0) |
Note that map's result type depends on the type of function that's passed to it. If the result type of that function argument is again an Int, the result of map is a BitSet, but if the result type of the function argument is something else, the result of map is just a Set. You'll find out soon how this type-flexibility is achieved in Scala.
The problem with BitSet is not an isolated case. Here are two more interactions with the interpreter that both map a function over a map:
| scala> Map("a" -> 1, "b" -> 2) map { case (x, y) => (y, x) } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res3: scala.collection.immutable.Map[Int,java.lang.String] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| = Map(1 -> a, 2 -> b) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> Map("a" -> 1, "b" -> 2) map { case (x, y) => y } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res4: scala.collection.immutable.Iterable[Int] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| = List(1, 2) |
You might ask, why not restrict map so that it can always return the same kind of collection? For instance, on bit sets map could accept only Int-to-Int functions and on maps it could only accept pair-to-pair functions. Not only are such restrictions undesirable from an object-oriented modelling point of view, they are illegal because they would violate the Liskov substitution principle: A Map is an Iterable. So every operation that's legal on an Iterable must also be legal on a Map.
Scala solves this problem instead with overloading: not the simple form of overloading inherited by Java (that would not be flexible enough), but the more systematic form of overloading that's provided by implicit parameters.
| def map[B, That](p: Elem => B) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (implicit bf: CanBuildFrom[B, That, This]): That = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val b = bf(this) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| for (x <- this) b += f(x) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| b.result | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Listing 3 shows trait TraversableLike's implementation of map. It's quite similar to the implementation of filter shown in class TraversableLike in Listing 2. The principal difference is that where filter used the newBuilder method, which is abstract in class TraversableLike, map uses a builder factory that's passed as an additional implicit parameter of type CanBuildFrom.
| package scala.collection.generic | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| trait CanBuildFrom[-From, -Elem, +To] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Creates a new builder | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(from: From): Builder[Elem, To] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Listing 4 shows the definition of the trait CanBuildFrom, which represents builder factories. It has three type parameters: Elem indicates the element type of the collection to be built, To indicates the type of collection to build, and From indicates the type for which this builder factory applies. By defining the right implicit definitions of builder factories, you can tailor the right typing behavior as needed. Take class BitSet as an example. Its companion object would contain a builder factory of type CanBuildFrom[BitSet, Int, BitSet]. This means that when operating on a BitSet you can construct another BitSet provided the type of the collection to build is Int. If this is not the case, you can always fall back to a different implicit builder factory, this time implemented in mutable.Set's companion object. The type of this more general builder factory, where A is a generic type parameter, is:
| CanBuildFrom[Set[_], A, Set[A]] |
So implicit resolution provides the correct static types for tricky collection operations such as map. But what about the dynamic types? Specifically, say you have a list value that has Iterable as its static type, and you map some function over that value:
| scala> val xs: Iterable[Int] = List(1, 2, 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| xs: Iterable[Int] = List(1, 2, 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> val ys = xs map (x => x * x) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ys: Iterable[Int] = List(1, 4, 9) |
Integrating new collections
What needs to be done if you want to integrate a new collection class, so that it can profit from all predefined operations at the right types? On the next few pages you'll be walked through two examples that do this.
Integrating sequences
| abstract class Base | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case object A extends Base | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case object T extends Base | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case object G extends Base | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case object U extends Base | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| object Base { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val fromInt: Int => Base = Array(A, T, G, U) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val toInt: Base => Int = Map(A -> 0, T -> 1, G -> 2, U -> 3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
| import collection.IndexedSeqLike | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import collection.mutable.{Builder, ArrayBuffer} | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import collection.generic.CanBuildFrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| final class RNA1 private (val groups: Array[Int], | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val length: Int) extends IndexedSeq[Base] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import RNA1._ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(idx: Int): Base = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| if (idx < 0 || length <= idx) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| throw new IndexOutOfBoundsException | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Base.fromInt(groups(idx / N) >> (idx % N * S) & M) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| object RNA1 { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Number of bits necessary to represent group | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val S = 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Number of groups that fit in an Int | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val N = 32 / S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Bitmask to isolate a group | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val M = (1 << S) - 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def fromSeq(buf: Seq[Base]): RNA1 = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val groups = new Array[Int]((buf.length + N - 1) / N) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| for (i <- 0 until buf.length) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| groups(i / N) |= Base.toInt(buf(i)) << (i % N * S) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new RNA1(groups, buf.length) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(bases: Base*) = fromSeq(bases) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Say you want to create a new sequence type for RNA strands, which are sequences of bases A (adenine), T (thymine), G (guanine), and U (uracil). The definitions for bases are easily set up as shown in Listing 5, RNA Bases.
Every base is defined as a case object that inherits from a common abstract class Base. The Base class has a companion object that defines two functions that map between bases and the integers 0 to 3. You can see in the examples two different ways to use collections to implement these functions. The toInt function is implemented as a Map from Base values to integers. The reverse function, fromInt, is implemented as an array. This makes use of the fact that both maps and arrays are functions because they inherit from the Function1 trait.
The next task is to define a class for strands of RNA. Conceptually, a strand of RNA is simply a Seq[Base]. However, RNA strands can get quite long, so it makes sense to invest some work in a compact representation. Because there are only four bases, a base can be identified with two bits, and you can therefore store sixteen bases as two-bit values in an integer. The idea, then, is to construct a specialized subclass of Seq[Base], which uses this packed representation.
Listing 6, RNA strands, presents the first version of this class. It will be refined later. The class RNA1 has a constructor that takes an array of Ints as its first argument. This array contains the packed RNA data, with sixteen bases in each element, except for the last array element, which might be partially filled. The second argument, length, specifies the total number of bases on the array (and in the sequence). Class RNA1 extends IndexedSeq[Base]. Trait IndexedSeq, which comes from package scala.collection.immutable, defines two abstract methods, length and apply. These need to be implemented in concrete subclasses. Class RNA1 implements length automatically by defining a parametric field of the same name. It implements the indexing method apply with the code given in class RNA1. Essentially, apply first extracts an integer value from the groups array, then extracts the correct two-bit number from that integer using right shift (>>) and mask (&). The private constants S, N, and M come from the RNA1 companion object. S specifies the size of each packet (i.e. two); N specifies the number of two-bit packets per integer; and M is a bit mask that isolates the lowest S bits in a word.
Note that the constructor of class RNA1 is private. This means that clients cannot create RNA1 sequences by calling new, which makes sense, because it hides the representation of RNA1 sequences in terms of packed arrays from the user. If clients cannot see what the representation details of RNA sequences are, it becomes possible to change these representation details at any point in the future without affecting client code. In other words, this design achieves a good decoupling of the interface of RNA sequences and its implementation. However, if constructing an RNA sequence with new is impossible, there must be some other way to create new RNA sequences, else the whole class would be rather useless. In fact there are two alternatives for RNA sequence creation, both provided by the RNA1 companion object. The first way is method fromSeq, which converts a given sequence of bases (i.e., a value of type Seq[Base]) into an instance of class RNA1. The fromSeq method does this by packing all the bases contained in its argument sequence into an array, then calling RNA1's private constructor with that array and the length of the original sequence as arguments. This makes use of the fact that a private constructor of a class is visible in the class's companion object.
The second way to create an RNA1 value is provided by the apply method in the RNA1 object. It takes a variable number of Base arguments and simply forwards them as a sequence to fromSeq. Here are the two creation schemes in action:
| scala> val xs = List(A, G, T, A) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| xs: List[Product with Base] = List(A, G, T, A) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> RNA1.fromSeq(xs) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res1: RNA1 = RNA1(A, G, T, A) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> val rna1 = RNA1(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| rna1: RNA1 = RNA1(A, U, G, G, T) |
Adapting the result type of RNA methods
Here are some more interactions with the RNA1 abstraction:
| scala> rna1.length | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res2: Int = 5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna1.last | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res3: Base = T | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna1.take(3) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res4: IndexedSeq[Base] = Vector(A, U, G) |
| final class RNA2 private ( | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val groups: Array[Int], | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val length: Int | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ) extends IndexedSeq[Base] with IndexedSeqLike[Base, RNA2] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import RNA2._ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override def newBuilder: Builder[Base, RNA2] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new ArrayBuffer[Base] mapResult fromSeq | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(idx: Int): Base = // as before | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Now that you understand why things are the way they are, the next question should be what needs to be done to change them? One way to do this would be to override the take method in class RNA1, maybe like this:
| def take(count: Int): RNA1 = RNA1.fromSeq(super.take(count)) |
To be able to do this, IndexedSeqLike bases itself on the newBuilder abstraction, which creates a builder of the right kind. Subclasses of trait IndexedSeqLike have to override newBuilder to return collections of their own kind. In class RNA2, the newBuilder method returns a builder of type Builder[Base, RNA2].
To construct this builder, it first creates an ArrayBuffer, which itself is a Builder[Base, ArrayBuffer]. It then transforms the ArrayBuffer builder by calling its mapResult method to an RNA2 builder. The mapResult method expects a transformation function from ArrayBuffer to RNA2 as its parameter. The function given is simply RNA2.fromSeq, which converts an arbitrary base sequence to an RNA2 value (recall that an array buffer is a kind of sequence, so RNA2.fromSeq can be applied to it).
If you had left out the newBuilder definition, you would have gotten an error message like the following:
| RNA2.scala:5: error: overriding method newBuilder in trait | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| TraversableLike of type => scala.collection.mutable.Builder[Base,RNA2]; | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| method newBuilder in trait GenericTraversableTemplate of type | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| => scala.collection.mutable.Builder[Base,IndexedSeq[Base]] has | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| incompatible type | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| class RNA2 private (val groups: Array[Int], val length: Int) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ^ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| one error found |
With the refined implementation of the RNA2 class, methods like take, drop, or filter work now as expected:
| scala> val rna2 = RNA2(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| rna2: RNA2 = RNA2(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna2 take 3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res5: RNA2 = RNA2(A, U, G) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna2 filter (U !=) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res6: RNA2 = RNA2(A, G, G, T) |
Dealing with map and friends
However, there is another class of methods in collections that are not dealt with yet. These methods do not always return the collection type exactly. They might return the same kind of collection, but with a different element type. The classical example of this is the map method. If s is a Seq[Int], and f is a function from Int to String, then s.map(f) would return a Seq[String]. So the element type changes between the receiver and the result, but the kind of collection stays the same.
There are a number of other methods that behave like map. For some of them you would expect this (e.g., flatMap, collect), but for others you might not. For instance, the append method, ++, also might return a result of different type as its arguments--appending a list of String to a list of Int would give a list of Any. How should these methods be adapted to RNA strands? Ideally we'd expect that mapping bases to bases over an RNA strand would yield again an RNA strand:
| scala> val rna = RNA(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| rna: RNA = RNA(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna map { case A => T case b => b } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res7: RNA = RNA(T, U, G, G, T) |
| scala> rna ++ rna | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res8: RNA = RNA(A, U, G, G, T, A, U, G, G, T) |
| scala> rna map Base.toInt | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res2: IndexedSeq[Int] = Vector(0, 3, 2, 2, 1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna ++ List("missing", "data") | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res3: IndexedSeq[java.lang.Object] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Vector(A, U, G, G, T, missing, data) |
| scala> val rna2 = RNA2(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| rna2: RNA2 = RNA2(A, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna2 map { case A => T case b => b } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res0: IndexedSeq[Base] = Vector(T, U, G, G, T) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> rna2 ++ rna2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res1: IndexedSeq[Base] = Vector(A, U, G, G, T, A, U, G, G, T) |
| def map[B, That](f: A => B) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (implicit cbf: CanBuildFrom[Repr, B, That]): That |
How is the That type determined? In fact it is linked to the other types by an implicit parameter cbf, of type CanBuildFrom[Repr, B, That]. These CanBuildFrom implicits are defined by the individual collection classes. In essence, an implicit value of type CanBuildFrom[From, Elem, To] says: "Here is a way, given a collection of type From, to build with elements of type Elem a collection of type To."
| final class RNA private (val groups: Array[Int], val length: Int) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| extends IndexedSeq[Base] with IndexedSeqLike[Base, RNA] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import RNA._ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Mandatory re-implementation of `newBuilder` in `IndexedSeq` | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override protected[this] def newBuilder: Builder[Base, RNA] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| RNA.newBuilder | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Mandatory implementation of `apply` in `IndexedSeq` | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(idx: Int): Base = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| if (idx < 0 || length <= idx) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| throw new IndexOutOfBoundsException | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Base.fromInt(groups(idx / N) >> (idx % N * S) & M) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // Optional re-implementation of foreach, | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| // to make it more efficient. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override def foreach[U](f: Base => U): Unit = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| var i = 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| var b = 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| while (i < length) { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| b = if (i % N == 0) groups(i / N) else b >>> S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| f(Base.fromInt(b & M)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| i += 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
| object RNA { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val S = 2 // number of bits in group | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val M = (1 << S) - 1 // bitmask to isolate a group | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| private val N = 32 / S // number of groups in an Int | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def fromSeq(buf: Seq[Base]): RNA = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val groups = new Array[Int]((buf.length + N - 1) / N) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| for (i <- 0 until buf.length) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| groups(i / N) |= Base.toInt(buf(i)) << (i % N * S) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new RNA(groups, buf.length) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(bases: Base*) = fromSeq(bases) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def newBuilder: Builder[Base, RNA] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new ArrayBuffer mapResult fromSeq | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| implicit def canBuildFrom: CanBuildFrom[RNA, Base, RNA] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new CanBuildFrom[RNA, Base, RNA] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(): Builder[Base, RNA] = newBuilder | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(from: RNA): Builder[Base, RNA] = newBuilder | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Now the behavior of map and ++ on RNA2 sequences becomes clearer. There is no CanBuildFrom instance that creates RNA2 sequences, so the next best available CanBuildFrom was found in the companion object of the inherited trait IndexedSeq. That implicit creates IndexedSeqs, and that's what you saw when applying map to rna2.
To address this shortcoming, you need to define an implicit instance of CanBuildFrom in the companion object of the RNA class. That instance should have type CanBuildFrom[RNA, Base, RNA]. Hence, this instance states that, given an RNA strand and a new element type Base, you can build another collection which is again an RNA strand. Listings 8 and 9 show the details. Compared to class RNA2, there are two important differences. First, the newBuilder implementation has moved from the RNA class to its companion object. The newBuilder method in class RNA simply forwards to this definition. Second, there is now an implicit CanBuildFrom value in object RNA. To create such an object you need to define two apply methods in the CanBuildFrom trait. Both create a new builder for an RNA collection, but they differ in their argument list. The apply() method simply creates a new builder of the right type. By contrast, the apply(from) method takes the original collection as argument. This can be useful to adapt the dynamic type of builder's return type to be the same as the dynamic type of the receiver. In the case of RNA this does not come into play because RNA is a final class, so any receiver of static type RNA also has RNA as its dynamic type. That's why apply(from) also simply calls newBuilder, ignoring its argument.
That is it. The final RNA class implements all collection methods at their natural types. Its implementation requires a little bit of protocol. In essence, you need to know where to put the newBuilder factories and the canBuildFrom implicits. On the plus side, with relatively little code you get a large number of methods automatically defined. Also, if you don't intend to do bulk operations like take, drop, map, or ++ on your collection you can choose to not go the extra length and stop at the implementation shown in for class RNA1.
The discussion so far centered on the minimal amount of definitions needed to define new sequences with methods that obey certain types. But in practice you might also want to add new functionality to your sequences or to override existing methods for better efficiency. An example of this is the overridden foreach method in class RNA. foreach is an important method in its own right because it implements loops over collections. Furthermore, many other collection methods are implemented in terms of foreach. So it makes sense to invest some effort optimizing the method's implementation. The standard implementation of foreach in IndexedSeq will simply select every i'th element of the collection using apply, where i ranges from 0 to the collection's length minus one. So this standard implementation selects an array element and unpacks a base from it once for every element in an RNA strand. The overriding foreach in class RNA is smarter than that. For every selected array element it immediately applies the given function to all bases contained in it. So the effort for array selection and bit unpacking is much reduced.
Integrating new sets and maps
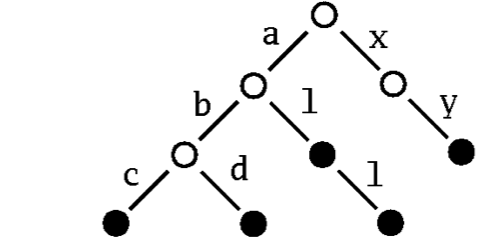
As a second example you'll learn how to integrate a new kind of map into the collection framework. The idea is to implement a mutable map with String as the type of keys by a "Patricia trie" The term Patricia is in fact an abbreviation for "Practical Algorithm to Retrieve Information Coded in Alphanumeric." The idea is to store a set or a map as a tree where subsequent character in a search key determines uniquely a descendant tree. For instance a Patricia trie storing the three strings "abc", "abd", "al", "all", "xy" would look like this:
To find the node corresponding to the string "abc" in this trie, simply follow the subtree labeled "a", proceed from there to the subtree labelled "b", to finally reach its subtree labelled "c". If the Patricia trie is used as a map, the value that's associated with a key is stored in the nodes that can be reached by the key. If it is a set, you simply store a marker saying that the node is present in the set.
| import collection._ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| class PrefixMap[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| extends mutable.Map[String, T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| with mutable.MapLike[String, T, PrefixMap[T]] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| var suffixes: immutable.Map[Char, PrefixMap[T]] = Map.empty | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| var value: Option[T] = None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def get(s: String): Option[T] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| if (s.isEmpty) value | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| else suffixes get (s(0)) flatMap (_.get(s substring 1)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def withPrefix(s: String): PrefixMap[T] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| if (s.isEmpty) this | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| else { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val leading = s(0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| suffixes get leading match { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case None => | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| suffixes = suffixes + (leading -> empty) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| case _ => | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| suffixes(leading) withPrefix (s substring 1) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override def update(s: String, elem: T) = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| withPrefix(s).value = Some(elem) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override def remove(s: String): Option[T] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| if (s.isEmpty) { val prev = value; value = None; prev } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| else suffixes get (s(0)) flatMap (_.remove(s substring 1)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def iterator: Iterator[(String, T)] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (for (v <- value.iterator) yield ("", v)) ++ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (for ((chr, m) <- suffixes.iterator; | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (s, v) <- m.iterator) yield (chr +: s, v)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def += (kv: (String, T)): this.type = { update(kv._1, kv._2); this } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def -= (s: String): this.type = { remove(s); this } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| override def empty = new PrefixMap[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Patricia tries support very efficient lookups and updates. Another nice feature is that they support selecting a subcollection by giving a prefix. For instance, in Figure 1 you can obtain the sub-collection of all keys that start with an "a" simply by following the "a" link from the root of the tree.
Based on these ideas we will now walk you through the implementation of a map that's implemented as a Patricia trie. We call the map a PrefixMap, which means that it provides a method withPrefix that selects a submap of all keys starting with a given prefix. We'll first define a prefix map with the keys shown in the running example:
| scala> val m = PrefixMap("abc" -> 0, "abd" -> 1, "al" -> 2, | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| "all" -> 3, "xy" -> 4) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| m: PrefixMap[Int] = Map((abc,0), (abd,1), (al,2), (all,3), | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| (xy,4)) |
| scala> m withPrefix "a" | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res14: PrefixMap[Int] = Map((bc,0), (bd,1), (l,2), (ll,3)) |
A prefix map node has two mutable fields: suffixes and value. The value field contains an optional value that's associated with the node. It is initialized to None. The suffixes field contains a map from characters to PrefixMap values. It is initialized to the empty map.
You might ask why did we pick an immutable map as the implementation type for suffixes? Would not a mutable map have been more standard, since PrefixMap as a whole is also mutable? The answer is that immutable maps that contain only a few elements are very efficient in both space and execution time. For instance, maps that contain fewer than 5 elements are represented as a single object. By contrast, the standard mutable map is a HashMap, which typically occupies around 80 bytes, even if it is empty. So if small collections are common, it's better to pick immutable over mutable. In the case of Patricia tries, we'd expect that most nodes except the ones at the very top of the tree would contain only a few successors. So storing these successors in an immutable map is likely to be more efficient.
Now have a look at the first method that needs to be implemented for a map: get. The algorithm is as follows: To get the value associated with the empty string in a prefix map, simply select the optional value stored in the root of the tree. Otherwise, if the key string is not empty, try to select the submap corresponding to the first character of the string. If that yields a map, follow up by looking up the remainder of the key string after its first character in that map. If the selection fails, the key is not stored in the map, so return with None. The combined selection over an option value is elegantly expressed using flatMap. When applied to an optional value, ov, and a closure, f, which in turn returns an optional value, ov flatMap f will succeed if both ov and f return a defined value. Otherwise ov flatMap f will return None.
The next two methods to implement for a mutable map are += and -=. In the implementation of PrefixMap, these are defined in terms of two other methods: update and remove.
The remove method is very similar to get, except that before returning any associated value, the field containing that value is set to None. The update method first calls withPrefix to navigate to the tree node that needs to be updated, then sets the value field of that node to the given value. The withPrefix method navigates through the tree, creating sub-maps as necessary if some prefix of characters is not yet contained as a path in the tree.
The last abstract method to implement for a mutable map is iterator. This method needs to produce an iterator that yields all key/value pairs stored in the map. For any given prefix map this iterator is composed of the following parts: First, if the map contains a defined value, Some(x), in the value field at its root, then ("", x) is the first element returned from the iterator. Furthermore, the iterator needs to traverse the iterators of all submaps stored in the suffixes field, but it needs to add a character in front of every key string returned by those iterators. More precisely, if m is the submap reached from the root through a character chr, and (s, v) is an element returned from m.iterator, then the root's iterator will return (chr +: s, v) instead. This logic is implemented quite concisely as a concatenation of two for expressions in the implementation of the iterator method in PrefixMap. The first for expression iterates over value.iterator. This makes use of the fact that Option values define an iterator method that returns either no element, if the option value is None, or exactly one element x, if the option value is Some(x).
| import scala.collection.mutable.{Builder, MapBuilder} | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| import scala.collection.generic.CanBuildFrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| object PrefixMap extends { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def empty[T] = new PrefixMap[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply[T](kvs: (String, T)*): PrefixMap[T] = { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| val m: PrefixMap[T] = empty | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| for (kv <- kvs) m += kv | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| m | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def newBuilder[T]: Builder[(String, T), PrefixMap[T]] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new MapBuilder[String, T, PrefixMap[T]](empty) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| implicit def canBuildFrom[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| : CanBuildFrom[PrefixMap[_], (String, T), PrefixMap[T]] = | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| new CanBuildFrom[PrefixMap[_], (String, T), PrefixMap[T]] { | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply(from: PrefixMap[_]) = newBuilder[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| def apply() = newBuilder[T] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| } |
Note that there is no newBuilder method defined in PrefixMap. There is no need to, because maps and sets come with default builders, which are instances of class MapBuilder. For a mutable map the default builder starts with an empty map and then adds successive elements using the map's += method. Mutable sets behave the same. The default builders for immutable maps and sets use the non-destructive element addition method +, instead of method +=.
However, in all these cases, to build the right kind of set or map, you need to start with an empty set or map of this kind. This is provided by the empty method, which is the last method defined in PrefixMap . This method simply returns a fresh PrefixMap.
We'll now turn to the companion object PrefixMap. In fact it is not strictly necessary to define this companion object, as class PrefixMap can stand well on its own. The main purpose of object PrefixMap is to define some convenience factory methods. It also defines a CanBuildFrom implicit to make typing work out better.
The two convenience methods are empty and apply. The same methods are present for all other collections in Scala's collection framework so it makes sense to define them here, too. With the two methods, you can write PrefixMap literals like you do for any other collection:
| scala> PrefixMap("hello" -> 5, "hi" -> 2) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res0: PrefixMap[Int] = Map((hello,5), (hi,2)) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| scala> PrefixMap.empty[String] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res2: PrefixMap[String] = Map() |
| scala> res0 map { case (k, v) => (k + "!", "x" * v) } | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| res8: PrefixMap[String] = Map((hello!,xxxxx), (hi!,xx)) |
Summary
To summarize, if you want to fully integrate a new collection class into the framework you need to pay attention to the following points:- Decide whether the collection should be mutable or immutable.
- Pick the right base traits for the collection.
- Inherit from the right implementation trait to implement most collection operations.
- If you want map and similar operations to return instances of your collection type, provide an implicit CanBuildFrom in your class's companion object.
You have now seen how Scala's collections are built and how you can build new kinds of collections. Because of Scala's rich support for abstraction, each new collection type can have a large number of methods without having to reimplement them all over again.
Resources
 |
Martin Odersky and Lex Spoon are coauthors of Programming in Scala, now available in a Second Edition: http://www.artima.com/shop/programming_in_scala_2ed |
The Scala programming language website is at:
http://www.scala-lang.org
The Scala 2.8 release notes are at:
http://www.scala-lang.org/node/7009
How to use Scala collections is described at:
http://lampwww.epfl.ch/~odersky/whatsnew/collections-api/collections.html
Talk back!
Have an opinion? Readers have already posted 6 comments about this article. Why not add yours?
About the authors
Lex Spoon is a software engineer at LogicBlox, Inc. He worked on Scala for two years as a post-doc at EPFL. He has a Ph.D. in computer science from Georgia Tech, where he worked on static analysis of dynamic languages. In addition to Scala, he has worked on a wide variety of programming languages, ranging from the dynamic language Smalltalk to the scientific language X10 to the logic language that powers LogicBlox. He and his wife currently live in Atlanta with two cats, a chihuahua, and a turtle.
Martin Odersky is the creator of the Scala language. As a professor at EPFL in Lausanne, Switzerland, he works on programming languages, more specifically languages for object-oriented and functional programming. His research thesis is that the two paradigms are two sides of the same coin, to be unified as much as possible. To prove this, he has experimented with a number of language designs, from Pizza to GJ to Functional Nets. He has also influenced the development of Java as a co-designer of Java generics and as the original author of the current javac reference compiler. Since 2001 he has concentrated on designing, implementing, and refining the Scala programming language.

Artima provides consulting and training services to help you make the most of Scala, reactive
and functional programming, enterprise systems, big data, and testing.
2070 N Broadway Unit 305
Walnut Creek CA 94597
USA
(925) 918-1769 (Phone)